Pre-lecture materials
Read ahead
Before class, you can prepare by reading the following materials:
Acknowledgements
Material for this lecture was borrowed and adopted from
Learning objectives
At the end of this lesson you will:
- Know the 5 basic built-in data types (or classes) of objects in R
- Know the types of attributes (or metadata) R objects can have
- Be able to create a vector, matrix, data frame, factor, and list in R
- Recognize missing values in R
R Nuts and Bolts
Entering Input
At the R prompt we type expressions. The <- symbol is the assignment operator.
The grammar of the language determines whether an expression is complete or not.
x <- ## Incomplete expressionThe # character indicates a comment. Anything to the right of the # (including the # itself) is ignored. This is the only comment character in R. Unlike some other languages, R does not support multi-line comments or comment blocks.
Evaluation
When a complete expression is entered at the prompt, it is evaluated and the result of the evaluated expression is returned. The result may be auto-printed.
The [1] shown in the output indicates that x is a vector and 5 is its first element.
Typically with interactive work, we do not explicitly print objects with the print function; it is much easier to just auto-print them by typing the name of the object and hitting return/enter. However, when writing scripts, functions, or longer programs, there is sometimes a need to explicitly print objects because auto-printing does not work in those settings.
When an R vector is printed you will notice that an index for the vector is printed in square brackets [] on the side. For example, see this integer sequence of length 20.
x <- 11:30
x
[1] 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30The numbers in the square brackets are not part of the vector itself, they are merely part of the printed output.
With R, it’s important that one understand that there is a difference between the actual R object and the manner in which that R object is printed to the console. Often, the printed output may have additional bells and whistles to make the output more friendly to the users. However, these bells and whistles are not inherently part of the object.
Note that the : operator is used to create integer sequences.
R Objects
R has five basic or “atomic” classes of vector objects:
logical:
FALSE,TRUE, andNAnumeric: real numbers
integer (and doubles): these are known collectively as numeric vectors
complex: complex numbers
character: the most complex type of atomic vector, because each element of a character vector is a string, and a string can contain an arbitrary amount of data.
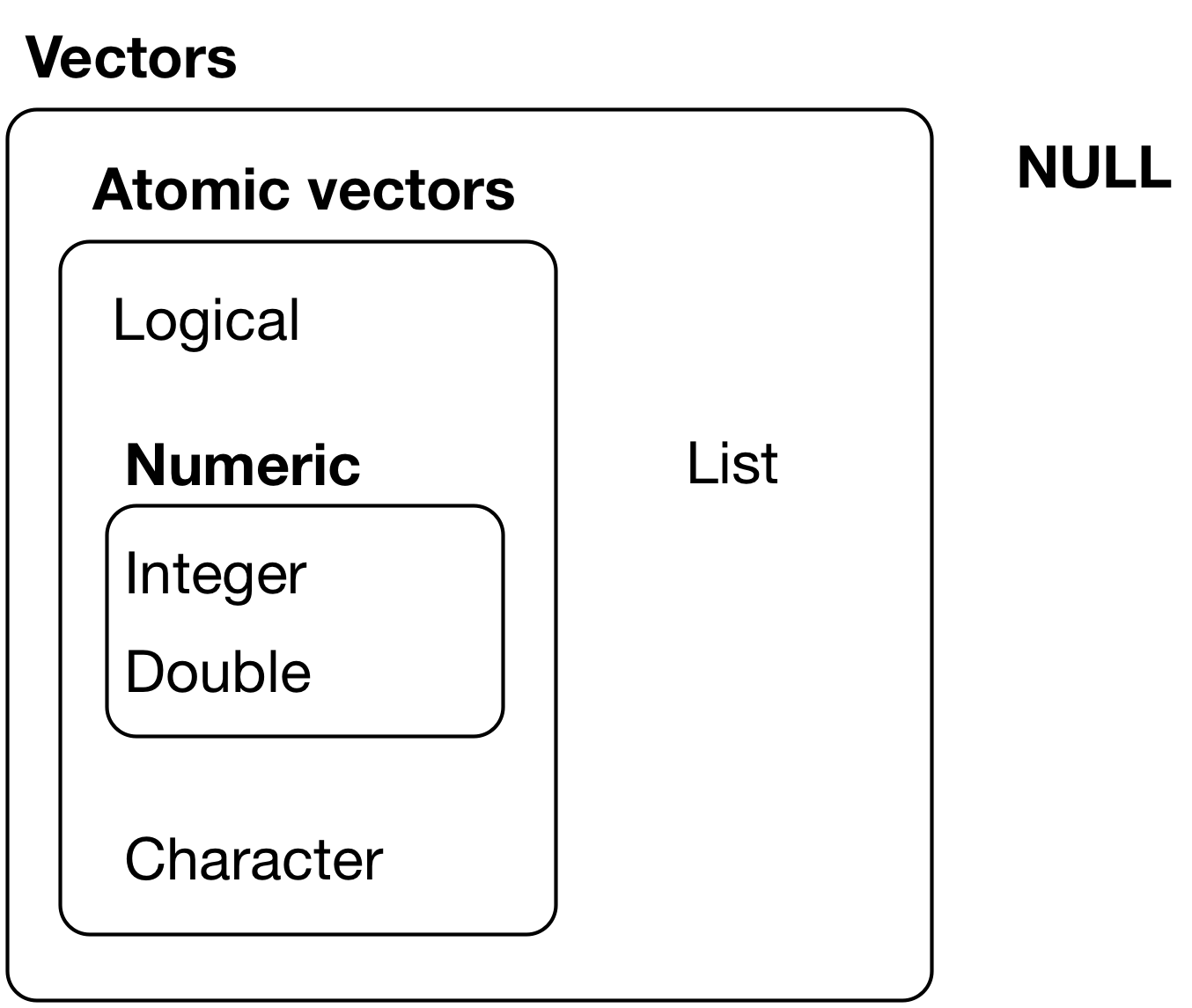
Figure 1: The hierarchy of R’s vector types
[Source: R 4 Data Science]
Integer and double vectors are known collectively as numeric vectors. In R, numbers are doubles by default. To make an integer, place an L after the number:
The distinction between integers and doubles is not usually important, but there are two important differences that you should be aware of:
Doubles are approximations. Doubles represent floating point numbers that can not always be precisely represented with a fixed amount of memory. This means that you should consider all doubles to be approximations.
For example, what is square of the square root of two?
The most basic type of R object is a vector. Empty vectors can be created with the vector() function. There is really only one rule about vectors in R, which is that A vector can only contain objects of the same class.
For example, if you run this, what happens?
vector("a", 1)
But of course, like any good rule, there is an exception, which is a list, which we will get to a bit later. A list is represented as a vector but can contain objects of different classes. Indeed, that’s usually why we use them.
There is also a class for “raw” objects, but they are not commonly used directly in data analysis and I won’t cover them here.
Numbers
Numbers in R are generally treated as numeric objects (i.e. double precision real numbers). This means that even if you see a number like “1” or “2” in R, which you might think of as integers, they are likely represented behind the scenes as numeric objects (so something like “1.00” or “2.00”). This isn’t important most of the time…except when it is.
If you explicitly want an integer, you need to specify the L suffix. So entering 1 in R gives you a numeric object; entering 1L explicitly gives you an integer object.
There is also a special number Inf which represents infinity. This allows us to represent entities like 1 / 0. This way, Inf can be used in ordinary calculations; e.g. 1 / Inf is 0.
The value NaN represents an undefined value (“not a number”); e.g. 0 / 0; NaN can also be thought of as a missing value (more on that later)
Attributes
R objects can have attributes, which are like metadata for the object. These metadata can be very useful in that they help to describe the object. For example, column names on a data frame help to tell us what data are contained in each of the columns. Some examples of R object attributes are
names, dimnames
dimensions (e.g. matrices, arrays)
class (e.g. integer, numeric)
length
other user-defined attributes/metadata
Attributes of an object (if any) can be accessed using the attributes() function. Not all R objects contain attributes, in which case the attributes() function returns NULL.
However, every vector has two key properties:
- Its type, which you can determine with
typeof().
- Its length, which you can determine with
length().
Creating Vectors
The c() function can be used to create vectors of objects by concatenating things together.
Note that in the above example, T and F are short-hand ways to specify TRUE and FALSE. However, in general one should try to use the explicit TRUE and FALSE values when indicating logical values. The T and F values are primarily there for when you’re feeling lazy.
You can also use the vector() function to initialize vectors.
x <- vector("numeric", length = 10)
x
[1] 0 0 0 0 0 0 0 0 0 0Mixing Objects
There are occasions when different classes of R objects get mixed together. Sometimes this happens by accident but it can also happen on purpose. So what happens with the following code?
In each case above, we are mixing objects of two different classes in a vector. But remember that the only rule about vectors says this is not allowed. When different objects are mixed in a vector, coercion occurs so that every element in the vector is of the same class.
In the example above, we see the effect of implicit coercion. What R tries to do is find a way to represent all of the objects in the vector in a reasonable fashion. Sometimes this does exactly what you want and…sometimes not. For example, combining a numeric object with a character object will create a character vector, because numbers can usually be easily represented as strings.
Explicit Coercion
Objects can be explicitly coerced from one class to another using the as.* functions, if available.
x <- 0:6
class(x)
[1] "integer"as.numeric(x)
[1] 0 1 2 3 4 5 6as.logical(x)
[1] FALSE TRUE TRUE TRUE TRUE TRUE TRUEas.character(x)
[1] "0" "1" "2" "3" "4" "5" "6"Sometimes, R can’t figure out how to coerce an object and this can result in NAs being produced.
x <- c("a", "b", "c")
as.numeric(x)
[1] NA NA NAas.logical(x)
[1] NA NA NAas.complex(x)
[1] NA NA NAWhen nonsensical coercion takes place, you will usually get a warning from R.
Matrices
Matrices are vectors with a dimension attribute. The dimension attribute is itself an integer vector of length 2 (number of rows, number of columns)
m <- matrix(nrow = 2, ncol = 3)
m
[,1] [,2] [,3]
[1,] NA NA NA
[2,] NA NA NAdim(m)
[1] 2 3attributes(m)
$dim
[1] 2 3Matrices are constructed column-wise, so entries can be thought of starting in the “upper left” corner and running down the columns.
m <- matrix(1:6, nrow = 2, ncol = 3)
m
[,1] [,2] [,3]
[1,] 1 3 5
[2,] 2 4 6Matrices can also be created directly from vectors by adding a dimension attribute.
m <- 1:10
m
[1] 1 2 3 4 5 6 7 8 9 10 [,1] [,2] [,3] [,4] [,5]
[1,] 1 3 5 7 9
[2,] 2 4 6 8 10Matrices can be created by column-binding or row-binding with the cbind() and rbind() functions.
x <- 1:3
y <- 10:12
cbind(x, y)
x y
[1,] 1 10
[2,] 2 11
[3,] 3 12rbind(x, y)
[,1] [,2] [,3]
x 1 2 3
y 10 11 12Lists
Lists are a special type of vector that can contain elements of different classes. Lists are a very important data type in R and you should get to know them well. Lists, in combination with the various “apply” functions discussed later, make for a powerful combination.
Lists can be explicitly created using the list() function, which takes an arbitrary number of arguments.
x <- list(1, "a", TRUE, 1 + 4i)
x
[[1]]
[1] 1
[[2]]
[1] "a"
[[3]]
[1] TRUE
[[4]]
[1] 1+4iWe can also create an empty list of a prespecified length with the vector() function
x <- vector("list", length = 5)
x
[[1]]
NULL
[[2]]
NULL
[[3]]
NULL
[[4]]
NULL
[[5]]
NULLFactors
Factors are used to represent categorical data and can be unordered or ordered. One can think of a factor as an integer vector where each integer has a label. Factors are important in statistical modeling and are treated specially by modelling functions like lm() and glm().
Using factors with labels is better than using integers because factors are self-describing. Having a variable that has values “Yes” and “No” or “Smoker” and “Non-Smoker” is better than a variable that has values 1 and 2.
Factor objects can be created with the factor() function.
[1] yes yes no yes no
Levels: no yestable(x)
x
no yes
2 3 ## See the underlying representation of factor
unclass(x)
[1] 2 2 1 2 1
attr(,"levels")
[1] "no" "yes"Often factors will be automatically created for you when you read a dataset in using a function like read.table(). Those functions often default to creating factors when they encounter data that look like characters or strings.
The order of the levels of a factor can be set using the levels argument to factor(). This can be important in linear modelling because the first level is used as the baseline level.
[1] yes yes no yes no
Levels: no yes[1] yes yes no yes no
Levels: yes noMissing Values
Missing values are denoted by NA or NaN for q undefined mathematical operations.
is.na()is used to test objects if they areNAis.nan()is used to test forNaNNAvalues have a class also, so there are integerNA, characterNA, etc.A
NaNvalue is alsoNAbut the converse is not true
## Create a vector with NAs in it
x <- c(1, 2, NA, 10, 3)
## Return a logical vector indicating which elements are NA
is.na(x)
[1] FALSE FALSE TRUE FALSE FALSE## Return a logical vector indicating which elements are NaN
is.nan(x)
[1] FALSE FALSE FALSE FALSE FALSEData Frames
Data frames are used to store tabular data in R. They are an important type of object in R and are used in a variety of statistical modeling applications. Hadley Wickham’s package dplyr has an optimized set of functions designed to work efficiently with data frames.
Data frames are represented as a special type of list where every element of the list has to have the same length. Each element of the list can be thought of as a column and the length of each element of the list is the number of rows.
Unlike matrices, data frames can store different classes of objects in each column. Matrices must have every element be the same class (e.g. all integers or all numeric).
In addition to column names, indicating the names of the variables or predictors, data frames have a special attribute called row.names which indicate information about each row of the data frame.
Data frames are usually created by reading in a dataset using the read.table() or read.csv(). However, data frames can also be created explicitly with the data.frame() function or they can be coerced from other types of objects like lists.
Data frames can be converted to a matrix by calling data.matrix(). While it might seem that the as.matrix() function should be used to coerce a data frame to a matrix, almost always, what you want is the result of data.matrix().
x <- data.frame(foo = 1:4, bar = c(T, T, F, F))
x
foo bar
1 1 TRUE
2 2 TRUE
3 3 FALSE
4 4 FALSEnrow(x)
[1] 4ncol(x)
[1] 2Names
R objects can have names, which is very useful for writing readable code and self-describing objects. Here is an example of assigning names to an integer vector.
x <- 1:3
names(x)
NULL New York Seattle Los Angeles
1 2 3 names(x)
[1] "New York" "Seattle" "Los Angeles"Lists can also have names, which is often very useful.
x <- list("Los Angeles" = 1, Boston = 2, London = 3)
x
$`Los Angeles`
[1] 1
$Boston
[1] 2
$London
[1] 3names(x)
[1] "Los Angeles" "Boston" "London" Matrices can have both column and row names.
c d
a 1 3
b 2 4Column names and row names can be set separately using the colnames() and rownames() functions.
Note that for data frames, there is a separate function for setting the row names, the row.names() function. Also, data frames do not have column names, they just have names (like lists). So to set the column names of a data frame just use the names() function. Yes, I know its confusing. Here’s a quick summary:
| Object | Set column names | Set row names |
|---|---|---|
| data frame | names() |
row.names() |
| matrix | colnames() |
rownames() |
Summary
There are a variety of different builtin-data types in R. In this chapter we have reviewed the following
atomic classes: numeric, logical, character, integer, complex
vectors, lists
factors
missing values
data frames and matrices
All R objects can have attributes that help to describe what is in the object. Perhaps the most useful attribute is names, such as column and row names in a data frame, or simply names in a vector or list. Attributes like dimensions are also important as they can modify the behavior of objects, like turning a vector into a matrix.
Post-lecture materials
Final Questions
Here are some post-lecture questions to help you think about the material discussed.
Questions:
Describe the difference between is.finite(x) and !is.infinite(x).
A logical vector can take 3 possible values. How many possible values can an integer vector take? How many possible values can a double take? Use google to do some research.
What functions from the readr package allow you to turn a string into logical, integer, and double vector?
Try and make a tibble that has columns with different lengths. What happens?
Additional Resources